About
|
DrugSense is a software tool able to predict the drug(s) to which the patient’s tumour will be most sensitive to by analysing the transcriptional profile of the tumour sample. The input of DrugSense is the transcriptional profile of the tumour, while the output is a list of drugs ranked according to their predicted potency in the patient, with the most potent at the top. DrugSense is based on a data-driven model derived from large publicly available collections of gene expression profiles of 1,000 cancer cell lines, and of dose-response curves of these cell lines to over 400 drugs and research compounds. |
|
Computational drug prediction in hepatoblastoma by integrating pan-cancer transcriptomics with pharmacological response. Hepatology 2023. Abstract |
|
|
The following section provides guidelines on how to make better use of DrugSense. |
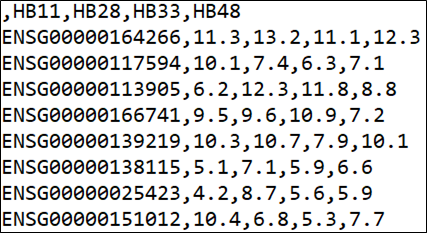
|
| Figure 1: DrugSense input file format. |
|
To get the best performance out of the algorithm, drug predictions for solid and liquid tumors are run separately. In this regard, it is recommended to pre-process data from solid and liquid tumors independently and to save the relative normalized expression profiles in separate files. In respect of data pre-processing, all the normalization methods suited for gene comparisons within a sample are well tolerated by the algorithm. In the following table are listed the most common one: |
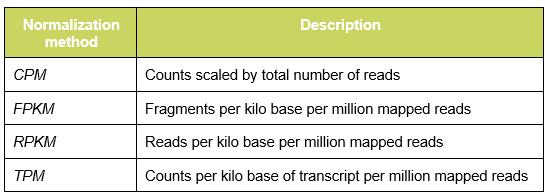
|
| Table 1: Common normalization method to account for gene comparisons within a sample. |
|
A specific drop-down menu (Figure 2) allows the user to select the tumor type corresponding to the expression profile(s) provided in input. This argument will instruct the algorithm on the drug-gene correlation profiles (DGCPs) to use; if no action is taken, the algorithm will import those trained on cancer cell lines from solid tumours, by default. |

|
| Figure 2: Drop-down menu to select the tumour type. |
|
|
|
Use of DrugSense tool is free to academic, government and non-profit users for non-commercial use. Use of DrugSense application for commercial use by non-profit and for-profit entities requires a license agreement. Parties interested in commercial use may first obtain a demo version of a stand-alone version of DrugSense application, subject to this License, by contacting Diego di Bernardo (dibernardo_at_tigem.it). Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
|
